Service: DNA sequencing – identification of biological material
Platforms
Fixlab
Techniques
Ancient dna analysis (adna)
Organization
Service contact persons
Phone:38612343170
DNA sequencing may be applied for the identification of a wide array of possible biological materials ranging from biodeteriogenic microorganisms (fungi/moulds and bacteria) to human, animal or plant materials. In the case of biodeteriogenic microorganisms, their identification can aid in cleaning and restoration, or may identify microorganisms, which are potentially dangerous to human health or are able to degrade the base substrate or painted layers. After DNA extraction, PCR amplification and DNA sequencing, the obtained DNA sequence is compared with the NCBI Blast database and thereby our biological material is identified.
In our laboratory at ZVKDS, DNA sequencing has been successfully applied for various cultural heritage items, which can be seen in our recent research publications: https://doi.org/10.1186/s40494-024-01527-4
https://doi.org/10.1007/s00248-024-02404-0
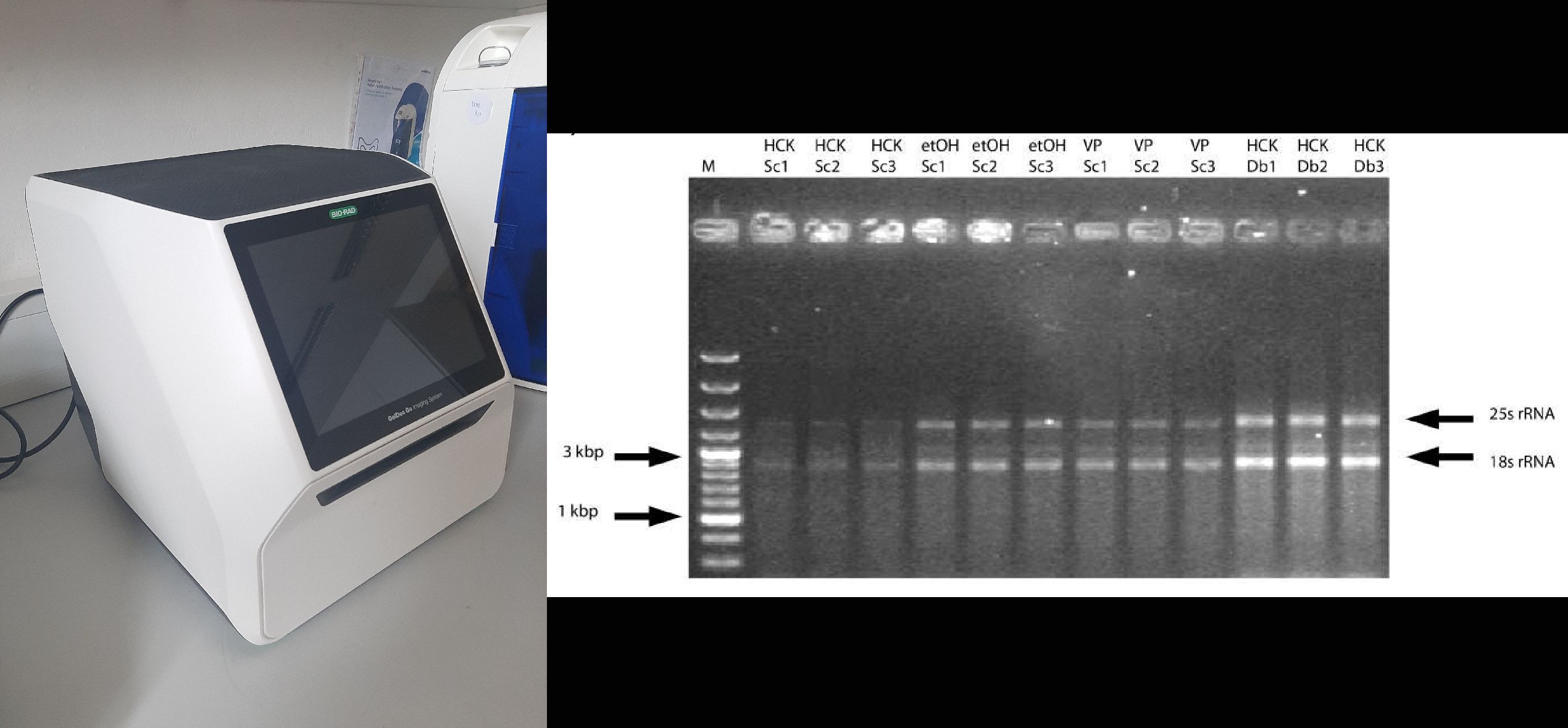
Apart from DNA sequencing, some biological materials may be identified using the GelDoc Go Gel Imaging System, which is based on agarose gel DNA electrophoresis imaging. This involves analyzing DNA bands generated after enzymatic restriction, where restriction enzymes cut the DNA molecule at specific sites, known as restriction recognition sites. These sites are determined by a specific sequence of 4–10 nucleotide bases in the DNA. Restriction enzymes detect polymorphisms among different biological species, as they cut the DNA molecule in a species-specific manner. Firstly the amplified DNA region (by PCR) is digested with restriction enzymes, and the resulting fragments are separated by agarose gel electrophoresis. The gel is visualized under fluorescence using a Gel Doc system, allowing for the identification of biological material based on the banding pattern.
-The analysis may take up to 3 months.
- Active biological material (microorganisms) has to be properly supplied (in PBS salyne buffer) within one week after swabbing.
Fields of application
Archaeology
Conservation (discipline)
Microbiology
Materials
Biological control agents
Biological material
Other information
-
Output: Research report with results and comments